The Pep Genie - Documentation
Table of Contents
Summary
The Pep Genie is a Django based website application that simplifies and speeds up the data analysis of peptide arrays.
Peptide arrays are small peptide libraries synthesised as a grid of spots on a membrane. Each spot in the array consists of a single unique peptide sequence. Binding of an overlaid target, such as a protein, is determined by immunodetection, and results in a grid of binding spot signals.
Peptide arrays are analysed by measuring spot intensity and slicing spot images (Figure 1). Done manually, this is a lengthy and painstaking process. The Pep Genie provides a grid drawing method for the quick measurement of peptide array binding spots and slicing of spot images, and prepares the data directly in PowerPoint presentation format, saving considerable of time and work. The Pep Genie is an open-source software that is freely available at the host website and the GitHub repository.
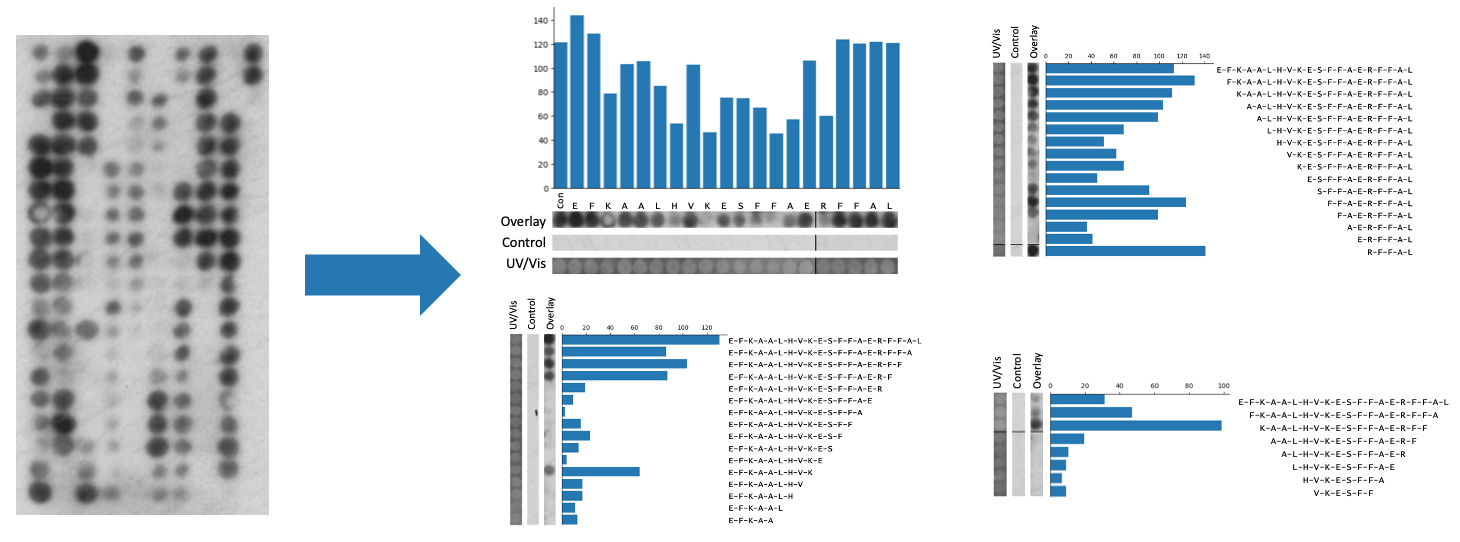
Figure 1: Peptide array analysis provided by The Pep Genie. Peptide array spots are quantified and sliced into figures.
http://pepgenie.pythonanywhere.com
https://github.com/thomas-wright-bioinformatics
Getting Started and Tutorials
Tutorials and a quick start guide are available here.
The Case for Automation
Peptide array analysis involves three main challenges.
-
Labour intensive densitometry measurement of tens to hundreds of binding spots
A typical peptide array experiment involves the analysis of 20-800 peptide binding spots. There is a lack of tools that allow for simple and rapid intensity measurement. Current options include the open-source software ImageJ/FIJI or Licor’s ImageStudio Lite.
A researcher would normally rely on freely available image analysis software such as the open-source software ImageJ/FIJI or Licor’s ImageStudio Lite. Measurement would entail drawing selection circles manually over each individual spot in the image. For large arrays consisting of hundreds of spots, this would be tedious and time-consuming. One tool, ImageStudio (full version), allows a researcher to draw a grid of spots without having to individually draw each spot. This is a considerable improvement, but the data is arranged in a difficult layout, and the software is available only by special request from Licor for groups who own a Licor Instrument. This is a restrictive requirement. The Pep Genie provides a grid drawing tool and automated analysis, allowing a researcher to quickly measure peptide spots.
-
Arrangement of binding spot ‘strips’ into a presentable figure
Perhaps the most time-consuming step in peptide array analysis is the preparation of the strips of spots for the final figure. In peptide array data, figures always include binding images of the columns of peptide spots (figure 2). This is analogous to the images of bands on a western blot figure. Three images are usually included: the test array, the control array, and the UV/Vis control array. These are positioned next to text labels that indicate their amino acid sequence. A bar graph may be included showing the intensity measurements of the spots. In standard peptide array experiments, there are usually three to ten of these figures, one for each peptide series of interest. For example, these can be an alanine scan and N,C-terminal truncations. To make these image slices/strips, a researcher would have to identify the correct spots from the grid of binding spots, and then screenshot the series of spots into a PowerPoint file. The strips must be of uniform thickness and perfectly aligned with each other. To add further to the challenge, a series of peptide spots can span more than one column on the peptide array. This means that a researcher must join together two screenshots of the spots, meanwhile making sure to maintain an even image width and proper alignment. Repeated 30 times, this makes for an arduous process. Quite often, these figures must be generated before it can be seen if the experiment was a technical success. If the experiment did not work, this work would have to be repeated. As part of the grid drawing tool of the Pep Genie, not only are the spots quickly measured, but images of the spots are automatically generated and joined together where needed. This automation saves considerable time.
-
Control spots can be too faint to align into a figure
In peptide array experiments, a blank image for the control array is desired, as this indicates a lack of non-specific binding. However, this makes it very difficult to make spot strip images and align them with the test and UV/Vis control images. If using the screenshotting method to capture the rows of spots or drawing circles to measure spots, it would be very hard to find them from a near-blank screen. As part of the grid drawing step in the Pep Genie, the grid for outlining the control array is automatically sized according to the previously aligned test array. With the width and height of the grid preset for the user, it is much easier to align the grid to the control spots.
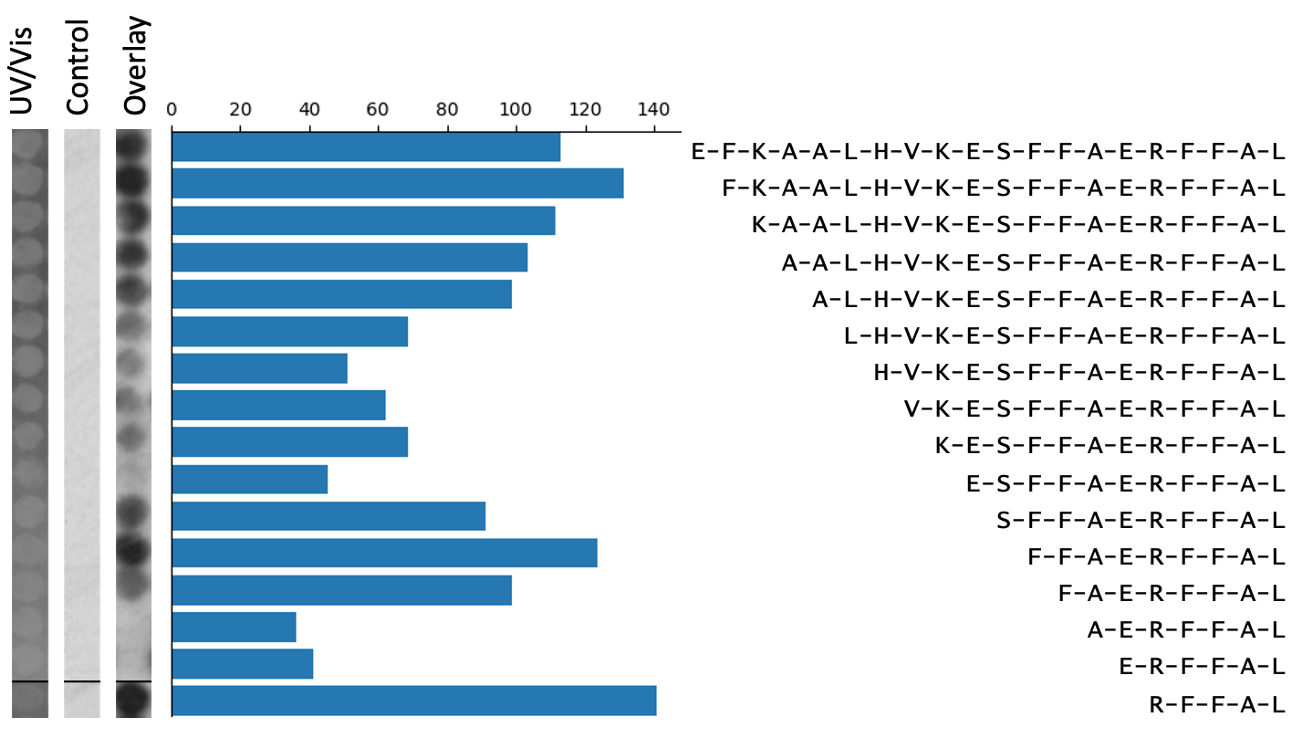
Figure 2: An example peptide array figure. Spot strip images must be perfectly aligned with each other, and with the graph and text.
Implementation
The core functionality of The Pep Genie is the grid drawing tool. This allows automation of the otherwise arduous tasks of spot quantification and image slicing. This feature has been expanded into a Django website application as follows:
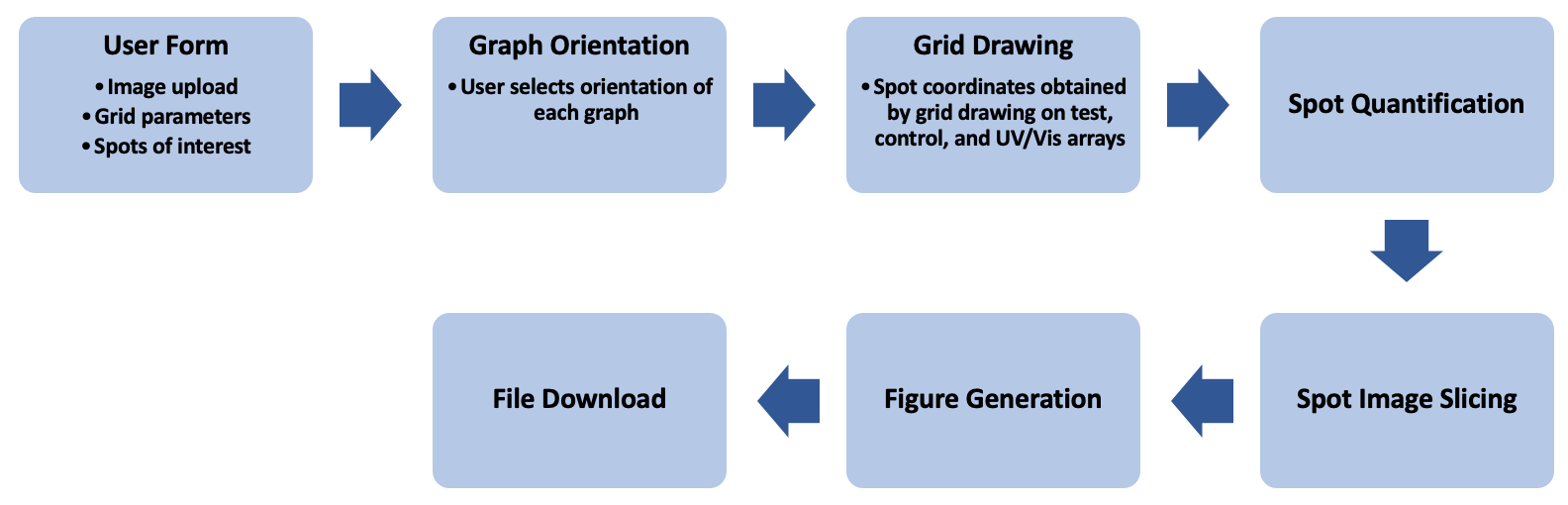
Figure 3: Workflow of The Pep Genie
-
Input of Image Files, Grid Parameters, and Spots of Interest by the User:
- The grid to be drawn is determined by the number of columns and number of the first column fields.
- The spots to be analysed in each figure is specified in the “Enter your Desired Strips” field.
-
Selection of graph orientation
- The user then selects the orientation of the final figures.
- Discovery arrays and truncation figures will have vertical spot slices and horizontal bar graphs.
- Alanine scans will have horizontal spot slices and a vertical bar graph.
-
Drawing and aligning the grid to the array images
- Based on the user-inputted description, a hidden html grid-div is generated with the required number of columns.
- Using JavaScript, the grid can be drawn by the user over their test, control and UV/Vis control array images.
- The pixel position of the grid on the image is passed to a hidden html form using JavaScript, and this information is passed to the Python backend.
-
Quantification of Binding Spots
- Each array image is cropped to the size of its drawn grid.
- Each spot is cropped into a separate image.
- The background is cropped out of each spot image using ImageOps.fit() and putalpha() functions from the Pillow package in Python.
- The mean pixel value is determined using the mean value from the ImageStat() function from the Pillow package.
- Values of the test array spots are subtracted from the control array spots. The raw and corrected values are included in the download zip file.
- UV/Vis spots are not quantified.
-
Slicing of Spot Images
- With each spot cropped into a separate image, spot images are joined end to end as specified by the user’s spots of interest.
- Series of spots that span multiple columns are joined in the same way, and a black line marks the junction. These strips are saved and included in the download file.
-
Arrangement of PowerPoint Figure Slides
- A PowerPoint file is assembled using the python-pptx library, and one slide is made for each figure.
- A bar plot is created from the corrected spot intensity data using the matplotlib.pyplot.bar() function from the Python library Matplotlib. The graph orientation is set by the user-inputted graph types. The graph is saved as a picture, then inserted into each slide.
- The spot slices for the test, control, and UV/Vis arrays are then inserted into the PowerPoint slides and are aligned with each other and to the bar chart.
-
Downloading the Data
- The figure PowerPoint file, the raw data, the corrected data, and the spot slice images are collected into a zip file using the Python Zipfile module. These files are then available for download by the user.
-
Manual Insertion of Sequence Labels
- The user can then insert the peptide sequence for each peptide spot manually.
- This text can be easily formatted to fit the spot and graph sizing. The figures are now finished.
-
What do I enter for “Number of Columns in Array”?
- This determines how many columns the blue grid will have when drawing the grid over your array images. This should therefore match the number of columns in your array. You can count the columns from the PEP file or check your UV/Vis control.
-
What do I enter for “Number of First Column”?
- This lets the application know how your array grid is numbered. This layout should match the “Enter your Desired Strips” field so that the application knows exactly which spots you want analyzed.
-
What do I enter for “Enter your Desired Strips”?
- This is where you tell the software which spots you want analyzed. You enter the first and last spots you want included in one graph and separate them by a dash. You then separate each graph by a comma. For example: “A03-R03, S03-G04”.
-
Why are my spot slices squint or cut off in my presentation file?
- The array images used may not be perfectly straight. It may be necessary to rotate the image and save it before using The Pep Genie. See the tutorial for more details on rotation.
- Drawing and aligning the grid to your images can be tricky and takes time. It may be worth trying again to get better alignment.
-
My UV/Vis is warped and will not align well in the figures.
- Because the UV/Vis control is captured on a camera, the image may be warped depending on the angle of the camera or if the array was not laid flat.
- It may be necessary to manually slice the spots from this image, making the slice wide enough to contain the warped columns.
- Alternatively, a new UV/Vis control can be taken using a UV/Vis light box.
-
The Application gives an error page when I click submit.
- Check that the information on the form page is correct, especially the “Enter your Desired Strips” field.
- Try navigating to the homepage and starting the workflow from the beginning.
-
The website is down, or the error page shows every time.
- Please post the issue on GitHub, and the problem will be addressed as soon as possible.
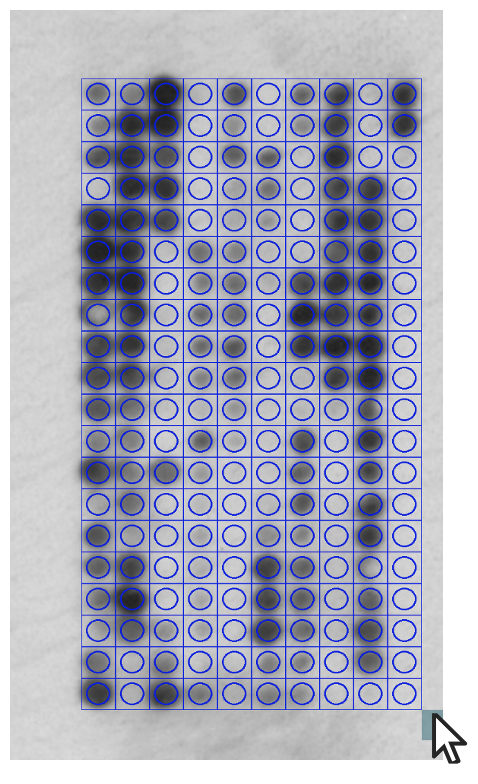
Figure 4: A grid being drawn over an array image.
FAQs
Useful References
Amartely, Hadar, Anat Iosub-Amir, and Assaf Friedler. "Identifying protein-protein interaction sites using peptide arrays." JoVE (Journal of Visualized Experiments) 93 (2014): e52097.
